PREGO Biobank
PREGO is a biobank of 5,707 DNA samples from blood donors whose four grandparents were born in the Bretagne and Pays-de-la-Loire regions of western France.
This unique resource enables studies in population genetics on the historical populations of western France, as well as genetic studies aimed at discovering rare, recent genetic variants associated with human disease.
The declaration and ethical approval process to set up PREGO was completed in 2013 and involved the Ministry of Research, the Committee for the Protection of Individuals (CNN in French), the Advisory Committee on Computing for Health Research (CCTIRS in French) and the National Commission on Computing and Freedom (CNIL in French).
Participants signed a written informed consent to participate in the study, to be included in the bioresource and to have their personal data processed.
The grandparents’ birthplaces for all PREGO participants are displayed below. 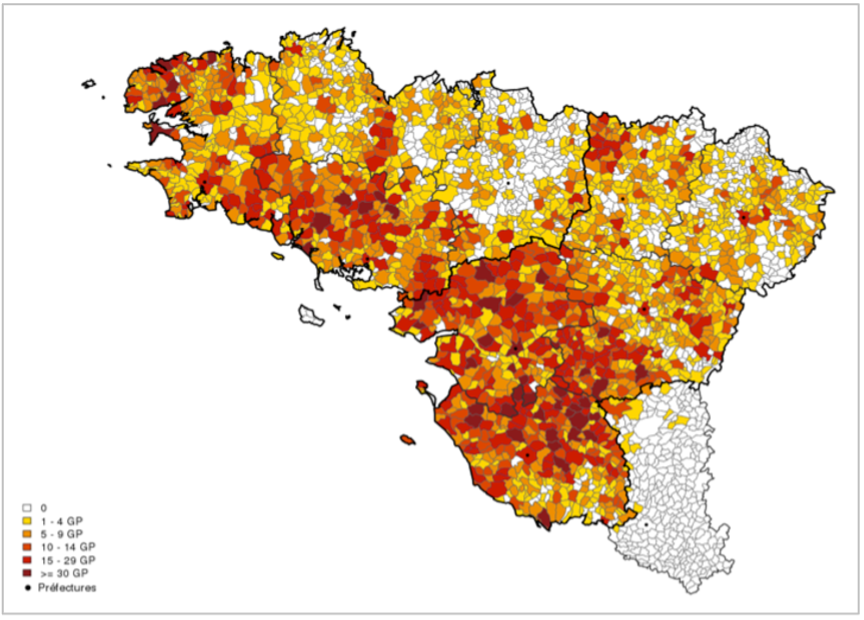
PREGO participants were recruited during 295 blood drives organised by the Etablissement Français du Sang (EFS) between February 2014 and March 2017, with an average of 19 donors per blood drive. Blood drives were sampled spatially and temporally to obtain as homogeneous a coverage as possible of the 9 departments included in the study. Priority was given to blood drives in rural areas.
The main inclusion criterion was that the 4 grandparents of each participant should preferably have been born within a radius of 30 km in western France.
Venous blood samples were taken for DNA extraction.
Participants completed a questionnaire on their grandparents', parents' and own birthplaces, place of residence, age, sex and information on previous participation in the study (by the individual or another family member).
No phenotypic or clinical data were collected at recruitment.
As a result, PREGO includes DNA from 83% of individuals whose 4 grandparents were born within 30 km of each other, and 66% within 15 km.
The mean age of participants is 49.7 years, with 56% of women (mean age: 47.5 years) and 44% of men (mean age: 51.4 years).
|
Max. distance between GPB* |
0 km |
< 5 km |
< 10 km |
< 15 km |
< 20 km |
< 25 km |
< 30 km |
< 40 km |
<50 km |
>=50 km |
N/A |
|
Cumulative % |
18% |
26% |
51% |
66% |
75% |
80% |
83% |
88% |
91% |
97% |
100% |
* GPB: grandparents’ birthplaces
Resource & Data Access
The PREGO biobank is hosted by the Centre de Ressources Biologiques of the CHU Nantes on behalf of l’institut du thorax.
PREGO array genotype data are accessible for academic research in compliance with INSERM policies, under the terms of a Data Access Agreement. The dataset consists of a core set of experimental genotypes at 210,171 autosomal sites measured on Affymetrix PMRA Axiom array plates and selected using the QC parameters described in Alves et al., for a group of 3,234 individuals selected from the PREGO biobank. The geographic spatial position (based on Lambert-93 projection) is also provided for each individual. The data are supplied in plink "bfile" format: the fam file indicates the coding and sex of the individual, the bim file indicates the SNPs and their chromosomal and physical positions, and the compressed bed file stores all the genotypes. Data access will be granted for not-for-profit use. Raw genotypes (on 920k markers, prior to QC) are also accessible, upon reasonable request. The data are also available from the European Genome-Phenome Archive (EGA) under accession number EGAD00010002661.
Any request regarding access to DNA samples or genotype data should be addressed to richard.redon@univ-nantes.fr.
Access to whole-genome sequencing data from the FranceGenRef study
The PREGO samples are also part of the FranceGenRef study, which aims to provide the community with a whole genome reference panel representative of the French historical population.
Participants were recruited on the basis of their grandparents' place of birth in order to describe the genetic diversity between French regions at the beginning of the 20th century. A total of 862 individuals originating from North-western France were sampled from three different studies: 354 blood donors from the PREGO biobank; 50 additional blood donors from the Finistère department; 458 individuals from the GAZEL cohort. All individuals signed an informed consent for genetic studies at the time of enrolment and blood sampling. Whole genome sequencing at 30x coverage was performed at the CNRGH. The resulting GVCF files were filtered to retain only SNPs with high mapping quality (MQ>30). Furthermore, only SNPs in Hardy-Weinberg equilibrium (HWE, p-value = 10-5) and with less than 10% missing data were retained.
In order to comply with French regulations on the protection of identifiable personal data, the individual genotype data will be submitted to the French Centralized Data Centre of the French Medicine Genomic Plan, which is under construction, and will then contribute to the Genome of Europe initiative. Pending these developments, requests for use of these data for research in population genetics should be directed to the LABEX GENMED. To access individual data, applicants should submit a synopsis of their project to the FrenchGenRef Data Access Committee composed of the three Principal Investigators*. Access will then be granted in compliance with the current French legislation on identifiable personal data and its compatibility of regulations with the requesting country.
* The Principal Investigators of the FranceGenRef study are:
Jean-François Deleuze ; Centre National de Recherche en Génomique Humaine, CEA, Evry, France
Emmanuelle Génin ; Inserm UMR1078, CHRU, Univ Brest, Brest, France
Richard Redon ; l’institut du thorax, Inserm UMR 1087/CNRS UMR 6291, Nantes Université, CHU de Nantes, Nantes, France
PREGO has already been a key resource for studies in biomedical, statistical or population genetics, as illustrated by the following publications:
2024
- Alves I, Giemza J, Blum MGB, Bernhardsson C, Chatel S, Karakachoff M, Saint Pierre A, Herzig AF, Olaso R, Monteil M, Gallien V, Cabot E, Svensson E, Bacq D, Baron E, Berthelier C, Besse C, Blanché H, Bocher O, Boland A, Bonnaud S, Charpentier E, Dandine-Roulland C, Férec C, Fruchet C, Lecointe S, Le Floch E, Ludwig TE, Marenne G, Meyer V, Quellery E, Racimo F, Rouault K, Sandron F, Schott J-J, Velo-Suarez L, Violleau J, Willerslev E, Coativy Y, Jézéquel M, Le Bris D, Nicolas C, Pailler Y, Goldberg M, Zins M, Le Marec H, Jakobsson M, Darlu P, Génin E, Deleuze J-F, Redon R, Dina C. Human genetic structure in Northwest France provides new insights into West European historical demography. Nat Commun 2024;15:6710.
- Herzig AF, Velo-Suárez L; FrEx Consortium; FranceGenRef Consortium; Dina C, Redon R, Deleuze JF, Génin E. How local reference panels improve imputation in French populations. Sci Rep. 2024 Jan 3;14(1):370. doi: 10.1038/s41598-023-49931-3.
2023
- Karam A, Delvallée C, Estrada-Cuzcano A, et al. WGS Revealed Novel BBS5 Pathogenic Variants, Missed by WES, Causing Ciliary Structure and Function Defects. Int J Mol Sci. 2023 May 13;24(10):8729. doi: 10.3390/ijms24108729.
- Odelin G, Faucherre A, Marchese D, et al. Variations in the poly-histidine repeat motif of HOXA1 contribute to bicuspid aortic valve in mouse and zebrafish. Nat Commun. 2023 Mar 20;14(1):1543. doi: 10.1038/s41467-023-37110-x.
- Ballinger ML, Pattnaik S, Mundra PA, et al. Heritable defects in telomere and mitotic function selectively predispose to sarcomas. Science. 2023 Jan 20;379(6629):253-260. doi: 10.1126/science.abj4784. Epub 2023 Jan 19.
2022
- Holstege H, Hulsman M, Charbonnier C, et al. Exome sequencing identifies rare damaging variants in ATP8B4 and ABCA1 as risk factors for Alzheimer's disease. Nat Genet. 2022 Dec;54(12):1786-1794. doi: 10.1038/s41588-022-01208-7. Epub 2022 Nov 21.
- Goudal A, Karakachoff M, Lindenbaum P, et al. Burden of rare variants in arrhythmogenic cardiomyopathy with right dominant form-associated genes provides new insights for molecular diagnosis and clinical management. Hum Mutat. 2022 Sep;43(9):1333-1342. doi: 10.1002/humu.24436. Epub 2022 Jul 23.
- Schramm C, Charbonnier C, Zaréa A, et al. Penetrance estimation of Alzheimer disease in SORL1 loss-of-function variant carriers using a family-based strategy and stratification by APOE genotypes. Genome Med. 2022 Jun 28;14(1):69. doi: 10.1186/s13073-022-01070-6.
- Herzig AF, Ciullo M; FranceGenRef Consortium; Leutenegger AL, Perdry H. Moment estimators of relatedness from low-depth whole-genome sequencing data. BMC Bioinformatics. 2022 Jun 24;23(1):254. doi: 10.1186/s12859-022-04795-8.
2021
- Rimbert A, Vanhoye X, Coulibaly D, et al. Phenotypic Differences Between Polygenic and Monogenic Hypobetalipoproteinemia. Arterioscler Thromb Vasc Biol. 2021 Jan;41(1):e63-e71.
- Delvallée C, Nicaise S, Antin M, et al. A BBS1 SVA F retrotransposon insertion is a frequent cause of Bardet-Biedl syndrome. Clin Genet. 2021 Feb;99(2):318-324. doi: 10.1111/cge.13878. Epub 2020 Nov 14.
- Quenez O, Cassinari K, Coutant S, et al. Detection of copy-number variations from NGS data using read depth information: a diagnostic performance evaluation. Eur J Hum Genet. 2021 Jan;29(1):99-109.
2020
- Belot A, Rice GI, Omarjee SO, et al. Contribution of rare and predicted pathogenic gene variants to childhood-onset lupus: a large, genetic panel analysis of British and French cohorts. Lancet Rheumatol. 2020 Feb;2(2):e99-e109.
- Bis JC, Jian X, Kunkle BW, et al. Whole exome sequencing study identifies novel rare and common Alzheimer's-Associated variants involved in immune response and transcriptional regulation. Mol Psychiatry. 2020 Aug;25(8):1859-1875.
2019
- Belbachir N, Portero V, Al Sayed ZR, et al. RRAD mutation causes electrical and cytoskeletal defects in cardiomyocytes derived from a familial case of Brugada syndrome. Eur Heart J. 2019 Oct 1;40(37):3081-3094.
- Kim A, Savary C, Dubourg C, et al. Integrated clinical and omics approach to rare diseases: novel genes and oligogenic inheritance in holoprosencephaly. Brain. 2019 Jan 1;142(1):35-49.
2018
- Vuillaume ML, Cogné B, Jeanne M, et al. Whole genome sequencing identifies a de novo 2.1 Mb balanced paracentric inversion disrupting FOXP1 and leading to severe intellectual disability. Clin Chim Acta. 2018 Oct;485:218-223.
- Bourcier R, Le Scouarnec S, Bonnaud S, et al. Rare Coding Variants in ANGPTL6 Are Associated with Familial Forms of Intracranial Aneurysm. Am J Hum Genet. 2018 Jan 4;102(1):133-141.
2017
- Bellenguez C, Charbonnier C, Grenier-Boley B, et al. Contribution to Alzheimer's disease risk of rare variants in TREM2, SORL1, and ABCA7 in 1779 cases and 1273 controls. Neurobiol Aging. 2017 Nov;59:220.e1-220.e9.
- Le Guennec K, Quenez O, Nicolas G, et al. 17q21.31 duplication causes prominent tau-related dementia with increased MAPT expression. Mol Psychiatry. 2017 Aug;22(8):1119-1125.
2016
- Colin E, Daniel J, Ziegler A, et al. Biallelic Variants in UBA5 Reveal that Disruption of the UFM1 Cascade Can Result in Early-Onset Encephalopathy. Am J Hum Genet. 2016 Sep 1;99(3):695-703.
- Le Guennec K, Nicolas G, Quenez O, et al. ABCA7 rare variants and Alzheimer disease risk. Neurology. 2016 Jun 7;86(23):2134-7.
- Nicolas G, Charbonnier C, Wallon D, et al. SORL1 rare variants: a major risk factor for familial early-onset Alzheimer's disease. Mol Psychiatry. 2016 Jun;21(6):831-6.